By Arron Carter
Sometimes, I feel like plant breeding is like the NFL draft. There are thousands of players out there, and, like an NFL scout, I walk around and look at their performance history and associated numbers.
In both scenarios, we find players (or in my case, breeding lines) that have a strong performance over the years. We then take a chance on them. NFL teams draft players hoping their performance in high school and college will translate into superior performance on the NFL team, and maybe even a future MVP. In my case, I hope performance over the multiple years of testing in various small plots trials throughout Washington will result in similar performance under full-scale commercial production.

What if an NFL scout had to make that selection without any previous knowledge? What if all you could do was look at a player in high school and decide if they would make it in the NFL. I am sure the accuracy of prediction would be very low. Looking just at a person on a high school football team and predicting they will be a future MVP of the NFL would be difficult. Many times, I do the same thing in my early generation plant breeding trials. I walk tens of thousands of rows, and basically, the only information I have is what the plant looks like. I can see when it flowers, how tall it is, how many tillers it has, how many seeds per head it has, but that is about it. Every now and then, we might get some stripe rust in the field, and I can evaluate for disease tolerance, but that does not happen every year, 2021 and 2023 being good examples. And then, it is very difficult to predict if a particular line has snow mold tolerance, good winter hardiness, good end-use quality, etc. All these traits are subsequently tested for over multiple years for evaluation. Each year, lines are discarded from the program (what we call negative selection) because they do not meet our standards. Eventually, one line rises to the top as having all the trait combinations and is released as the next cultivar. Our prediction accuracy during this process can be low. We start with thousands of lines in field testing, only to end up with one for commercial release. So, what if there was a way we could increase our prediction accuracy?
The field of genomics has been making major improvements over the past decade in wheat. We now have the first whole genome sequence of wheat. We have the capability of finding hundreds of DNA markers in wheat. As a reminder, a DNA marker is a region of the DNA that is associated with a specific trait of interest. We have the capabilities to identify polymorphisms, which are differences at the DNA level, between different lines of interest. For example, if you took the well-known cultivars Eltan and Madsen and sequenced them, you would find tens of thousands of polymorphisms between them — regions in their DNA that are different and make them unique cultivars.
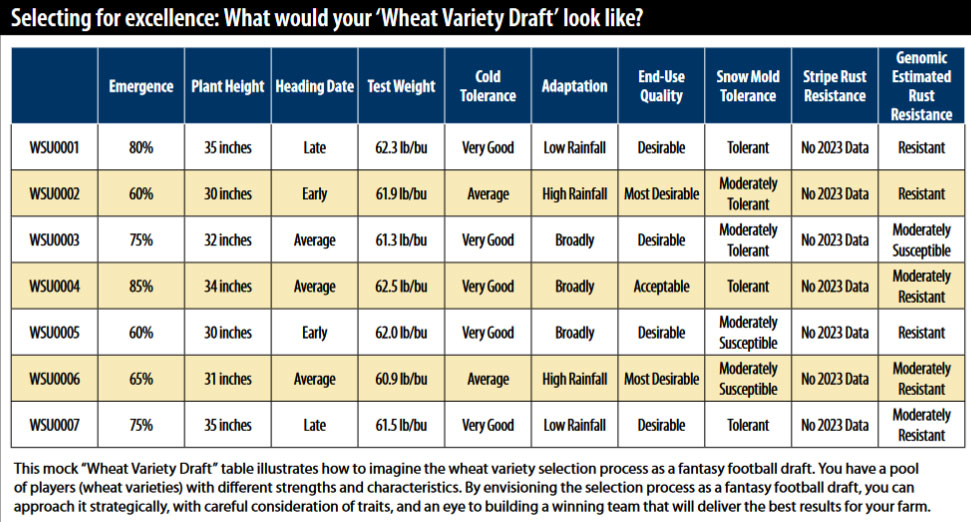
What does all this have to do with plant breeding, selection, and selection efficiency? For years, animal breeders have been using a technique called genomic selection. This is a process where selection of an animal to be used in mating is determined on their genetics, not on their phenotypic performance. With our improved knowledge of wheat genomics, we can now use this in wheat breeding.
To use genomic selection, you first gather up thousands of lines that have both genotypic and phenotypic data collected on them. In my breeding program, we use past breeding lines that we have already collected data on as part of the breeding process. A computer program is used to associate every DNA polymorphism (or difference) between the lines with the associated trait of interest. We then go to new lines that have only been genotyped but have no phenotype data associated with them yet. Using this computer program, we look at all the DNA polymorphisms in these new lines and find the lines with the highest number of polymorphisms that are associated with our favorable traits. The program returns an “estimated breeding value” that is an indication of how well the line is expected to perform for a certain trait given its genotype.

What is the value in all of this? When I now look at lines for the first time, not only can I see what they visually look like, but I have estimates of performance for traits I am interested in. If I look at a line and it visually looks appealing but has a low estimate for disease resistance or end-use quality, I can pass it over. This prevents us from spending valuable resources field testing lines that will not meet expectations. It also allows us to make estimates in years we cannot get data from the field. For example, in 2021 and 2023 when we did not have much disease pressure in the field, I could still make an estimate to assist with selection. Since we do not get end-use quality data in time for planting decisions, I can make estimates of quality and not replant anything that has a low estimate. It has proven to be a very useful tool in the breeding program.
We can use this process for many traits, although some are more useful in predicting than others. For traits with a little simpler genetic control, like end-use quality or disease resistance, the accuracy of prediction is very high. For more complex traits like grain yield, predictions can be lower. Regardless, the ability to have these estimated prediction values is proving more useful than having nothing at all. As more data is collected on different traits and more is understood about wheat genetics, this process will continue to improve.
Relying on both genetic and phenotypic data to make selections is becoming common in breeding programs. There will always be a need to test breeding lines under field conditions, but using genomic selection will assist in selecting and advancing lines that have the best chance of become the next great cultivar.
This article originally appeared in the July 2023 issue of Wheat Life Magazine.

Arron Carter
Arron Carter, Ph.D., holds the O.A. Vogel Endowed Chair of Wheat Breeding and Genetics at Washington State University. His research is directed toward breeding improved wheat varieties for cropping systems in Washington state that incorporate diverse rotations and environments. His research goal is to release high-yielding, disease resistant varieties with good end-use quality that will maintain profitability and reduce the risk to growers. New varieties are developed using a combination of traditional plant breeding methods, molecular marker technology, and biotechnology. Read more about Dr. Carter.
